Celiac disease, a chronic autoimmune disorder that affects about 1% of the global population, is most commonly managed by adhering to a strict gluten-free diet.
Gluten, found in wheat, barley, rye, and some oats, can cause significant intestinal damage to those with the condition. Recently, Dr. Veronica Dodero from Bielefeld University and her team have made a breakthrough in understanding how gluten exacerbates this condition.
The primary culprit in celiac disease is a gluten protein that, when consumed, is not fully broken down by the digestive system. This incomplete digestion can lead to larger fragments of gluten, known as peptides, lingering in the gut.
For individuals with active celiac disease, these peptides can induce what’s known as leaky gut syndrome, where the gut lining becomes more permeable than normal.
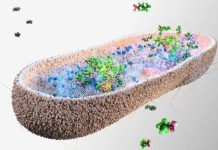
Dr. Dodero’s team focused on a specific gluten peptide called the 33-mer deamidated gliadin peptide (DGP). This peptide is particularly troublesome because it forms nanosized structures called oligomers that accumulate in the gut lining.
These structures can pry open the tightly closed gut lining, allowing harmful substances to seep into the bloodstream, which triggers inflammation and further complications.
This discovery is significant because it challenges the conventional understanding of how celiac disease progresses. Previously, it was believed that chronic inflammation from the disease led to a leaky gut.
However, the new findings suggest that the damage to the gut lining from DGP oligomers might actually be the initial trigger for the inflammation seen in celiac disease.
The study, which involved high-resolution microscopy and biophysical techniques, was conducted by an interdisciplinary team that included Dr. Maria Georgina Herrera from the University of Buenos Aires.
Their work revealed that when these DGP oligomers accumulate, they significantly increase the permeability of the gut lining in their test models.
Another layer to this issue is the role of human leukocyte antigens (HLAs), which are proteins on the surface of cells that help the immune system distinguish between the body’s own cells and foreign invaders.
In celiac disease, certain HLA proteins (HLA-DQ2 and HLA-DQ8) are involved. The structure of the 33-mer DGP fits perfectly into these HLAs, initiating an aggressive immune response that leads to inflammation and damage to the small intestine.
This fitting of DGP into HLA-DQ2 or HLA-DQ8 is so precise that it turns the DGP into what scientists call a superantigen, which is an antigen that triggers an excessive immune response.
Understanding this interaction is crucial as it underscores the importance of maintaining a gluten-free diet for those diagnosed with celiac disease, as this is currently the only effective treatment to prevent these reactions.
The implications of these findings are profound. They not only offer new insights into the biological mechanisms behind celiac disease but also pave the way for potentially new therapeutic approaches that could target these specific interactions within the gut.
Such treatments could offer new hope to those who suffer from this debilitating condition, providing them with more options beyond dietary restrictions.
The study, published in the journal Angewandte Chemie International Edition, marks a significant step forward in the scientific understanding of celiac disease and could lead to more effective ways of managing the condition in the future.
If you care about coffee, please read studies that drinking coffee this way can help prevent stroke, heart disease, and drink coffee after breakfast, not before, for better blood sugar control.
For more information about nutrition, please see recent studies about natural supplement that could relieve anxiety, and results showing this common food oil in the U.S. can change genes in the brain.
The research findings can be found in Angewandte Chemie International Edition.
Copyright © 2024 Knowridge Science Report. All rights reserved.