A UCLA research team has devised a technique that extends the capabilities of fluorescence microscopy, which allows scientists to precisely label parts of living cells and tissue with dyes that glow under special lighting.
The researchers use artificial intelligence to turn two-dimensional images into stacks of virtual three-dimensional slices showing activity inside organisms.
In a study published in Nature Methods, the scientists also reported that their framework, called “Deep-Z,” was able to fix errors or aberrations in images, such as when a sample is tilted or curved.
Further, they demonstrated that the system could take 2D images from one type of microscope and virtually create 3D images of the sample as if they were obtained by another, more advanced microscope.
“This is a very powerful new method that is enabled by deep learning to perform 3D imaging of live specimens, with the least exposure to light, which can be toxic to samples,” said senior author Aydogan Ozcan, UCLA chancellor’s professor of electrical and computer engineering and associate director of the California NanoSystems Institute at UCLA.
In addition to sparing specimens from potentially damaging doses of light, this system could offer biologists and life science researchers a new tool for 3D imaging that is simpler, faster and much less expensive than current methods.
The opportunity to correct for aberrations may allow scientists studying live organisms to collect data from images that otherwise would be unusable. Investigators could also gain virtual access to expensive and complicated equipment.
This research builds on an earlier technique Ozcan and his colleagues developed that allowed them to render 2D fluorescence microscope images in super-resolution.
Both techniques advance microscopy by relying upon deep learning — using data to “train” a neural network, a computer system inspired by the human brain.
Deep-Z was taught using experimental images from a scanning fluorescence microscope, which takes pictures focused at multiple depths to achieve 3D imaging of samples.
In thousands of training runs, the neural network learned how to take a 2D image and infer accurate 3D slices at different depths within a sample.
Then, the framework was tested blindly — fed with images that were not part of its training, with the virtual images compared to the actual 3D slices obtained from a scanning microscope, providing an excellent match.
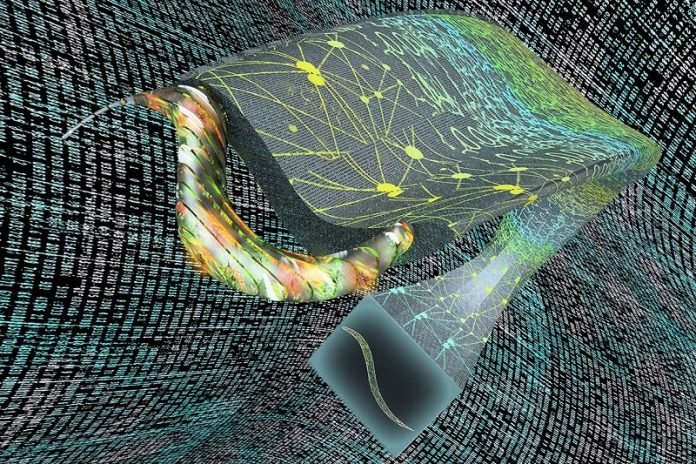
Ozcan and his colleagues applied Deep-Z to images of C. elegans, a roundworm that is a common model in neuroscience because of its simple and well-understood nervous system. Converting a 2D movie of a worm to 3D, frame by frame, the researchers were able to track the activity of individual neurons within the worm body.
And starting with one or two 2D images of C. elegans taken at different depths, Deep-Z produced virtual 3D images that allowed the team to identify individual neurons within the worm, matching a scanning microscope’s 3D output, except with much less light exposure to the living organism.
The researchers also found that Deep-Z could produce 3D images from 2D surfaces where samples were tilted or curved — even though the neural network was trained only with 3D slices that were perfectly parallel to the surface of the sample.
“This feature was actually very surprising,” said Yichen Wu, a UCLA graduate student who is co-first author of the publication. “With it, you can see through curvature or other complex topology that is very challenging to image.”
In other experiments, Deep-Z was trained with images from two types of fluorescence microscopes: wide-field, which exposes the entire sample to a light source; and confocal, which uses a laser to scan a sample part by part. Ozcan and his team showed that their framework could then use 2D wide-field microscope images of samples to produce 3D images nearly identical to ones taken with a confocal microscope.
This conversion is valuable because the confocal microscope creates images that are sharper, with more contrast, compared to the wide field. On the other hand, the wide-field microscope captures images at less expense and with fewer technical requirements.
“This is a platform that is generally applicable to various pairs of microscopes, not just the wide-field-to-confocal conversion,” said co-first author Yair Rivenson, UCLA assistant adjunct professor of electrical and computer engineering.
“Every microscope has its own advantages and disadvantages. With this framework, you can get the best of both worlds by using AI to connect different types of microscopes digitally.”
Other authors are graduate students Hongda Wang and Yilin Luo, postdoctoral fellow Eyal Ben-David and Laurent Bentolila, scientific director of the California NanoSystems Institute’s Advanced Light Microscopy and Spectroscopy Laboratory, all of UCLA; and Christian Pritz of Hebrew University of Jerusalem in Israel.
The research was supported by the Koç Group, the National Science Foundation and the Howard Hughes Medical Institute. Imaging was performed at CNSI’s Advanced Light Microscopy and Spectroscopy Laboratory and Leica Microsystems Center of Excellence.
Written by Wayne Lewis.