Scientists at Columbia have developed a gene-editing tool—using jumping genes—that inserts any DNA sequence into the genome without cutting, fixing a major shortcoming of existing CRISPR technology.
A new discovery could fix one of the major shortcomings of current gene-editing tools, including CRISPR, and offer a powerful new approach for genetic engineering and gene therapy.
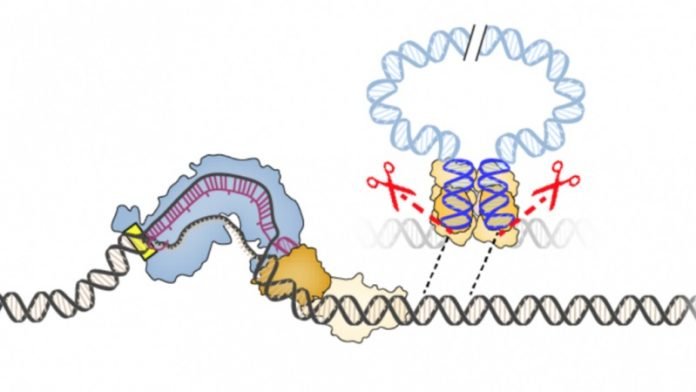
Their new technology, called INTEGRATE, harnesses bacterial jumping genes to reliably insert any DNA sequence into the genome without cutting DNA.
Current gene-editing tools rely on cutting DNA, but those cuts can lead to errors.
“Current tools are like molecular scissors: they cut DNA, but the actual editing is performed by the cell’s own DNA repair machinery,” says Sam Sternberg, PhD, assistant professor of biochemistry and molecular biophysics at Columbia and senior author of the new study.
“You’re at the mercy of the cell to finish the job.”
The new INTEGRATE technology, published online today in the journal Nature, functions more like molecular glue than molecular scissors.
“Rather than introduce DNA breaks and rely on the cell to repair the break, INTEGRATE directly inserts a user-defined DNA sequence at a precise location in the genome, a capability that molecular biologists have sought for decades,” says Sternberg, who was recently recruited to Columbia from Jennifer Doudna’s laboratory at UC Berkeley.
Current tools are finicky
Editing a cell’s genome with current tools is like using a word processor to edit a huge document, but with software that has a mind of its own.
Typically, researchers want to make a small change at one specific sequence of DNA bases, leaving the rest of the genome untouched.
The best current tool, built with components from one kind of bacterial CRISPR-Cas system, cuts both strands of the DNA molecule at a specific sequence, like adding a paragraph break to a block of text.
These breaks are only the starting point: the actual ‘editing’ is performed by the cell’s own DNA repair machinery, often using DNA sequences provided by the researcher to fill in the gap.
Relying on the cell’s repair machinery has major limitations. Many cells repair the DNA break incorrectly or introduce mistakes in the process, and other cells may not even express the necessary repair machinery to insert new genetic payloads.
In addition, DNA breaks trigger a DNA damage response that can have other adverse effects.
This makes gene editing difficult or impossible in some cell types, and severely constrains researchers’ ability to introduce precise genetic modifications in a safe manner.
INTEGRATE system harnesses jumping genes
The new work solves that problem with a self-contained DNA editing system – derived from Vibrio cholera bacteria — that doesn’t require any assistance from the cell.
CRISPR is a class of natural defense systems in bacteria that are remarkably diverse.
To find new gene-editing tools, Sternberg and three graduate students looked to bacteria to find variations of well-studied CRISPR-Cas systems with unusual properties that would reveal new tool capabilities.
This search led them to a transposon, or “jumping gene,” found in the bacterium Vibrio cholerae. This transposon has co-opted the bacterium’s CRISPR-Cas system, normally used to thwart mobile genetic elements, to insert itself into different regions of the bacterial genome.
Sternberg and his students found that the transposon integrates into specific sites in the bacterial genome, not by cutting DNA into two, but by using a separate enzyme to slip the transposon into the genome. Importantly, the site where the enzyme, an integrase, inserts the DNA is completely controlled by its associated CRISPR system.
The researchers harnessed this discovery to create a gene-editing tool that can be programed to insert any DNA sequence into any site in a bacterial genome. Like CRISPR, the integrase finds the appropriate site with a guide RNA.
By reprogramming the guide RNA, Sternberg and his students were able to precisely control the location where a piece of donor DNA was integrated. And by replacing the transposon sequence with other DNA payloads, they could insert sequences up to 10,000 bases long into a bacterial genome.
Thus, the INTEGRATE technology, unlike other editing tools based on integrases, is the first fully programmable insertion system studied to date.
Sequencing the edited bacteria confirmed that the payloads were inserted precisely, with no extra copies at off-target sites.
Improved gene editing
With INTEGRATE, a set of enzymes can carry out the entire DNA integration process, reliably inserting an arbitrary DNA payload into a precise location within a cell’s genome, without relying on the host cell’s DNA repair machinery.
The technique should enable a vast range of new gene editing opportunities. Many biotechnology products, including gene and cell therapies, engineered crops, and biologics, require precise integration of large genetic payloads.
The INTEGRATE technology offers a fresh new approach with the same programmability and ease of use as CRISPR-Cas9, but without the adverse effects associated with DNA breaks.
“We can program this CRISPR-transposon system to integrate its genetic payload at virtually any genomic site, and by understanding how it works, we will be able to engineer it to be even more effective,” says Sternberg.
Next Steps
Sternberg’s team developed the INTEGRATE technology using bacterial genetics experiments, and they are now testing it in additional cell types, including mammalian cells.
Based on the developmental trajectory of CRISPR-Cas9 technology, Sternberg says there’s good reason to believe that precise DNA integration will work as effectively in mammalian cells as it does in E. coli, opening up the door to basic research uses and eventual clinical applications.
DOI: https://doi.org/10.1038/s41586-019-1323-z.



